Transgene Activation with Antibiotics.
Transgene Activation with Antibiotics.
Several gene regulation systems for independent modulation of transgenic expression via antibiotics are available at PolyGene (e.g. macrolide inducibility, Eon; Weber et al, 2002).
Mode of Function
These unique systems, developed at the Swiss Federal Institute of Technology (ETH) in Zurich, Switzerland, are based on the capacity of antibiotics to bind repressor molecules and thus their capacity to modulate gene expression. Crossing of two complementary (EFFECTOR and RESPONDER) transgenic mouse lines yields offspring, in which expression of the Gene Of Interest (GOI) is actively inhibited.
Transcription is de-repressed by administration of the respective antibiotic, which dissociates the fused Operator-Binding Protein (OBP) from the Operator. GOI expression is thus unleashed and free to be driven by a definable (e.g. tissue specific) promoter.

Transcription is de-repressed by administration of the respective antibiotic, which dissociates the fused Operator-Binding Protein (OBP) from the Operator. GOI expression is thus unleashed and free to be driven by a definable (e.g. tissue specific) promoter.

The EFFECTOR mouse expresses an Operator Binding Protein (OBP) which can be linked to further functional proteins, e.g. to a trans-silencing (e.g. KRAB) domain as represented here (STOP). The RESPONDER mouse is engineered to harbour the GOI and a functional promoter (P), combined with operator (O) DNA sequences, to which the OBP has binding affinity.
Dose-Response
While binding of OBP alone strongly reduces GOI expression, it is fully abolished when KRAB-OBP fusion protein is bound to the operator sequences (Fussenegger et al., 2000).
1. Expression, as shown in cell culture experiments, is gradually increased up to roughly 100-fold at antibiotic concentrations ranging from about 0.1 to 2 μg/ml (Fussenegger et al., 2000, Weber et al., 2002).
2. As evidenced in vitro, protein production follows linear kinetics over a period of approx. 70 hours (Weber et al., 2002).
1. Expression, as shown in cell culture experiments, is gradually increased up to roughly 100-fold at antibiotic concentrations ranging from about 0.1 to 2 μg/ml (Fussenegger et al., 2000, Weber et al., 2002).
2. As evidenced in vitro, protein production follows linear kinetics over a period of approx. 70 hours (Weber et al., 2002).
Bioavailability
Macrolides (e.g. macrolide), many of which have had a proven track record as antibiotic agents for over half a century, are reputed for excellent bioavailability and optimal pharmacokinetics. The low concentrations required for adjusting responsive elements are reached in all tissues following as well intravenous as oral administration.
System Compatibility in vivo
Tested on mice with implanted HT-1080 cell derivatives, the Eon system was also proven to respond in vivo, in a dose-dependent manner (Weber et al., 2002).
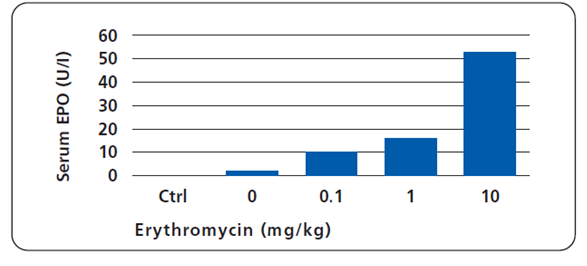

Microencapsulated cells producing erythropoietin (EPO) under macrolide control were implanted in the peritoneal cavity of receptor mice. EPO synthesis was monitored 72 hrs after injection of Macrolide at indicated doses. Non-transplanted control mice (Ctrl), irrespective of the quantities of antibiotic administered, showed endogenous EPO production levels at the detection limit.
Independent Control
As evidenced in antibiotic interference studies (Weber et al., 2002), the different systems are fully compatible, thus paving the way for independent regulation of different transgenes in a single cell or combinatorial transcription control (Kramer et al., 2002).
Positioning
PolyGene offers free promoter choice in RESPONDER constructs, thus ideally positioning Eon DEREPRESSOR systems for tissue specific applications. Note that the initial RESPONDER mouse, exhibiting promoter-driven GOI expression, can serve as constitutive EXPRESSOR.
Caution is however warranted for transgenes affecting embryogenesis. Hence, the DEREPRESSOR systems are best suited for transgene expression modulation in adult mice.
Streptogramins as well as macrolides are known for their low interference with host metabolism. Apart from possible concerns in the field of immunology or in that of neurology the antibiotic ON systems are indicated for research applications in a broad range of therapeutic areas.
Caution is however warranted for transgenes affecting embryogenesis. Hence, the DEREPRESSOR systems are best suited for transgene expression modulation in adult mice.
Streptogramins as well as macrolides are known for their low interference with host metabolism. Apart from possible concerns in the field of immunology or in that of neurology the antibiotic ON systems are indicated for research applications in a broad range of therapeutic areas.
References
Fussenegger, M. et al.,
Streptogramin-based gene regulation systems for mammalian cells.
Nat. Biotechnol. 18, 1203-1208 (2000)< Weber, W. et al.,
Macrolide-based transgene control in mammalian cells and mice.
Nat. Biotechnol.20, 901-906 (2002)
Streptogramin-based gene regulation systems for mammalian cells.
Nat. Biotechnol. 18, 1203-1208 (2000)< Weber, W. et al.,
Macrolide-based transgene control in mammalian cells and mice.
Nat. Biotechnol.20, 901-906 (2002)
Kramer, B. et al.,
BioLogic Gates Enable Logical Transcription Control in Mammalian Cells.
Biotechnol. Bioeng.87, 478-484 (2002)



