Simple and rapid design of genetically engineered rodent models.
Customized mouse and rat model generation via the nuclease-based system CRISPR/Cas9 outcompetes traditional methods in terms of time. Additionally, CRISPRs can be designed to target virtually all genes in a eukaryotic genome, just by synthesis of short sequence specific RNAs.
Like TALENs, the CRISPR/Cas9 system (for review see Doudna and Charpentier, 2014 and Hsu et al., 2014 ) is used to introduce double strand breaks into a genome in a sequence specific manner. Whereas TALENs use a combination of hard-to-assemble protein domains recognizing the specific DNA sequence, the CRISPR/Cas9 system makes use of easily synthesized small RNA molecules that are applicable to a wider veriety of target sequences. These small RNAs, originally derived from Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) in bacteria, target the nuclease Cas9 to the specific genomic region and introduce double strand breaks (Jinek et al., 2012 ).
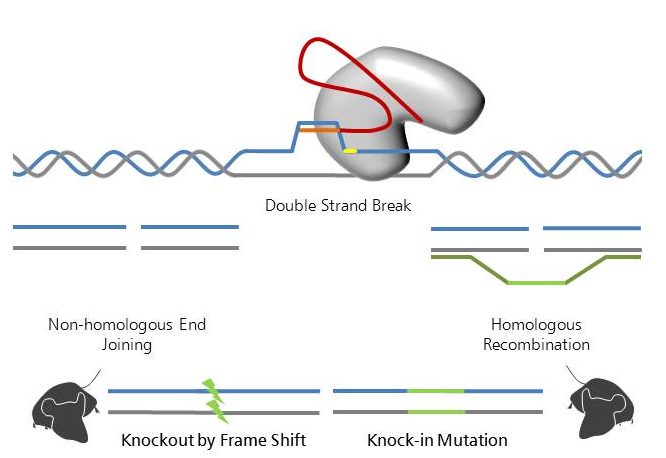
As compared to TALENs and other nuclease based techniques, CRISPR/Cas9 is the perfect system in terms of cutting efficiency. However, as the higher efficiency may lead to a higher number of off-target cutting for CRISPR/Cas9, a thorough analysis of off-targets is highly recommended.
CRISPR/Cas9 rodent model generation possibilities at PolyGene:
CRISPRs Service Details GET A QUOTE





